
NRI researchers significantly expand the toolkit for fruit fly researchers
 To tease out function of a gene, researchers need to do some sleuthing to find out some of its basic biological characteristics such as which tissues it is expressed during various developmental stages, subcellular localization of protein it encodes and how loss or disruption of the gene or its protein product affects physiology and/or morphology at the cellular or organismal levels.
To tease out function of a gene, researchers need to do some sleuthing to find out some of its basic biological characteristics such as which tissues it is expressed during various developmental stages, subcellular localization of protein it encodes and how loss or disruption of the gene or its protein product affects physiology and/or morphology at the cellular or organismal levels.
To address these questions, traditionally a research lab would obtain or generate numerous reagents such as loss-of-function mutants, RNAi lines to knockdown genes, a specific antibody against each protein, and others. While highly informative, this approach of generating reagents for each gene one-by-one is expensive, laborious and time-consuming for individual labs. For instance, despite decades of research, antibodies are currently only available for about ~3-4% of all protein coding genes in fruit flies.
This week, a paper published in the journal eLife by a collaborative team of researchers headed by Drs. Hugo J Bellen, professor at the Baylor College of Medicine and HHMI investigator at the Neurological Research Institute at Texas Children’s Hospital, Allan Spradling, HHMI investigator at the Carnegie Institution for Science, and Roger Hoskins at the Lawrence Berkeley National Laboratory describes the use of Minos transposon-based MiMIC vector to create a very valuable resource of more than 7,500 lines with insertions at different positions in the Drosophila genome.
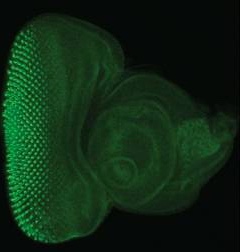
While all types of insertions are valuable, Minos Mediated Integration Cassette (MiMIC) insertions in coding introns, when inserted in the proper orientation, create gene trap (truncating) mutations that can be used to study loss-of-function phenotypes. MiMIC act as highly efficient and reliable gene traps; 92% of gene trap MiMIC insertions in essential genes were lethal whereas none of the MiMICs inserted in the non-gene trap orientation were lethal. Importantly, the MiMIC insertion cassette within coding introns can be converted into an artificial exon that encodes green fluorescent protein (GFP) through recombination.
In this study, the authors used this method to generate a collection of more than 400 EGFP tagged genes that will be made publicly available through the Bloomington Stock Center and can be accessed via a public searchable database.
During the course of this large-scale undertaking, they discovered that approximately 75% of genes with internal GFP tags retain normal function and roughly 90% had high enough expression to be detected in live tissues without fixation. This resource of endogenously, internally tagged proteins obviates the need to generate protein specific antibodies by providing reliable, robust information about cellular and subcellular protein localization.
Furthermore, they demonstrated that using a combination of genetic tools, these tagged genes/proteins can be spatially, temporally, and reversibly knocked-down. This means consequences of loss-of-function of a tagged gene/protein can be examined at specific developmental stages, within specific tissues, that will go a long way in understanding how a particular gene/protein functions.
In summary, authors have generated an extremely valuable genetic resource that permits unprecedented manipulations in live animals and will allow researchers to address many important yet previously inaccessible, questions.