An international team discovers a novel Alzheimer’s disease risk gene among Icelanders
 Drs. Shinya Yamamoto, Joshua M. Shulman, Hugo J. Bellen, faculty at Baylor College of Medicine and researchers in the Neurological Research Institute (NRI) at Texas Children’s Hospital are part of a multi-center collaborative study that identified a novel genetic risk factor for late-onset Alzheimer’s disease (LOAD). Postdoctoral fellow, Dr. David Li-Kroeger, and graduate student, Jose Salazar, also contributed to the study published in the journal PLoS Genetics this week.
Drs. Shinya Yamamoto, Joshua M. Shulman, Hugo J. Bellen, faculty at Baylor College of Medicine and researchers in the Neurological Research Institute (NRI) at Texas Children’s Hospital are part of a multi-center collaborative study that identified a novel genetic risk factor for late-onset Alzheimer’s disease (LOAD). Postdoctoral fellow, Dr. David Li-Kroeger, and graduate student, Jose Salazar, also contributed to the study published in the journal PLoS Genetics this week.
Alzheimer’s disease (AD) is the most common cause of dementia among older adults and the presence of certain genetic variants increases an individual’s risk for developing AD. In recent years, methodologic advances have accelerated the discovery of genetic variants responsible for early- and late-onset AD; however, confirming the functional impact of identified variants and their role in disease pathogenesis remains a major challenge.
Identifying rare genetic variants of AD helps to unravel novel cellular mechanisms that can cause AD. Depending on a population’s migration pattern and history, rare variants are frequently enriched in certain ethnic or geographic populations. For instance, a protective APP variant occurs predominantly only in Scandinavian populations whereas AKAP9 variants are relatively common among African-Americans.
In this study, an exome-wide genotyping array was employed to discover rare genetic variants that cause LOAD. Exomes of 1393 subjects with LOAD were compared with the exomes of 8141 neurologically healthy control individuals.
The consortium found a missense variant in TM2D3, a gene that has never been studied in human or other vertebrate species. Interestingly, while the probability of this variant was very rare among people of European ancestry, it was significantly enriched among Icelanders (but still 1% frequency). Carriers of this variant were estimated to have an approximately 6-fold increased risk of developing AD.
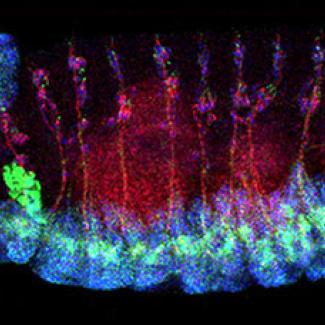
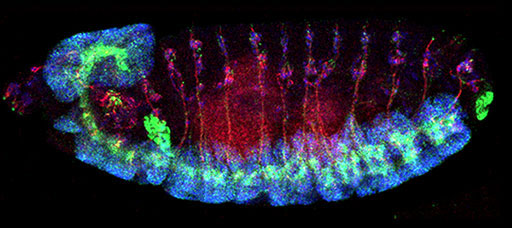
To further understand the functional impact of the discovered mutation and its potential link to AD mechanisms, the NRI team under the leadership of Yamamoto, Shulman and Bellen undertook functional analysis of TM2D3 in the fruit fly model system.
They found that the TM2D3 gene is conserved from fruit flies to man and its loss-of-function causes abnormal development of the nervous system. They next demonstrated that human TM2D3 could functionally substitute for its fly counterpart, and rescue the neurodevelopmental defect; whereas, introduction of the AD-associated variant was unable to restore the normal function.
This research illustrates how experiments performed in simple model organisms can complement human genetic discoveries. First, it shows that the fruit fly and human versions of TM2D3 are functionally interchangeable. Second, since the discovered variant has a significant functional impact in flies, it follows that this genetic variant may be similarly damaging in humans. Lastly, a potential role for TM2D3 in AD pathogenesis is suggested, and the team is now investigating the molecular mechanisms.
In summary, this study has identified a novel, rare genetic variant in the TM2D3 gene to be a risk factor in the development of late-onset Alzheimer’s disease among Icelanders. This gene has not been previously linked to Alzheimer’s, and researchers are hopeful that further investigations may lead to new and more effective therapies.